GNBF5010 Homework 2¶
Student id: 1155228903
Question1 (Unknown Letters)¶
The input file seqs.txt contains 516 sequences, with each sequence in a single line. Write a program to list which letters in the file seqs.txt are not A, T, C, or G. It should only list each letter once.
def main():
with open("seqs.txt", 'r') as file:
seqs = file.read()
non_ATCG_letters = find_non_ATCG_letters(seqs)
print("Letters in sequences that are not A, T, C, or G:")
print(' '.join(non_ATCG_letters))
# Find which letters in these sequences are not A, T, C, or G. It should only list each letter once.
def find_non_ATCG_letters(seqs):
non_ATCG = set()
for seq in seqs.splitlines():
for letter in seq:
if letter not in 'ATCG':
non_ATCG.add(letter)
return non_ATCG
if __name__ == "__main__":
main()
Letters in sequences that are not A, T, C, or G: M K S W N R H Y
Question2 (Sequence Properties)¶
Write a program, 1) read all sequences in seqs.txt and store them into a list called seqs, 2) prompt the user a menu for selection of various properties of the sequences, and 3) show the corresponding results based on user’s choice. 4) Prompt the selection menu again and exit the program only if the user enters ‘0’; otherwise, repeat steps 2 and 3.
Your program should keep prompting the selection menu until the user inputs “0”. The menu includes the following items. Please use separate functions in your program for the processes in options 1 to 4.
- Number of sequences in the input file
- The total number of occurrences of a short sequence (e.g. GGATC) in all sequences in the input file. The program will then prompt another message for the input of the target short sequence. Note that no need to consider overlapping cases. For example, the number of occurrences of AAA in AAAAAA is two.
- Number of sequences with length ≥ a particular length, e.g. 500. The program will then ask for the minimum length.
- Number of sequences with GC content ≥ a given value, e.g. 50%. The GC content could be calculated as (num_of_G + num_of_C) / Total_num_of_AGCT.
- The combination of choices 3 and 4: Number of sequences with length ≥ a particular length and with GC content ≥ a particular value.
def main():
with open("seqs.txt", 'r') as file:
seqs = [line.strip() for line in file if line.strip()]
while True:
print("===========================")
print("Please select the sequences property that you want to display or press 0 to exit the program.")
print("1) Total number of sequences")
print("2) Number of pattern occurrences")
print("3) Number of sequences with length >= min_len")
print("4) Number of sequences with GC% >= min_GC")
print("5) Number of sequences with length >= min_len and GC% >= min_GC\n")
choice = input("Enter the choice: ")
if choice == '0':
print("Exiting...")
break
elif choice == '1':
print(f"Total number of sequences is {count_total_sequences(seqs)}.\n")
elif choice == '2':
pattern = input("Enter the short sequence to search for (e.g. GGATC): ")
print(f"Number of occurrences of '{pattern}' is {count_pattern_occurrences(seqs, pattern)}.\n")
elif choice == '3':
min_len = int(input("min_len = "))
print(f"Number of sequences with length >= min_len is {count_sequences_by_length(seqs, min_len)}.\n")
elif choice == '4':
min_gc = int(input("min_GC (e.g. 50%) = "))
print(f"Num of seqs with gc >= min_gc is {count_sequences_by_gc_content(seqs, min_gc)}.\n")
elif choice == '5':
min_len = int(input("min_len = "))
min_gc = int(input("min_GC (e.g. 50%) = "))
print(f"Number of seqs with len >= min_len and gc >= min_gc is {count_sequences_by_length_and_gc(seqs, min_len, min_gc)}.\n")
else:
print("Invalid choice, please try again.")
# 1) Number of sequences in the input file.
def count_total_sequences(seqs):
return len(seqs)
# 2) The total number of occurrences of a short sequence (e.g. GGATC) in all sequences in the input file.
def count_pattern_occurrences(seqs, pattern):
count = 0
for seq in seqs:
if pattern in seq:
count += seq.count(pattern)
return count
# 3) Number of sequences with length ≥ a particular length, e.g. 500. The program will then ask for the minimum length.
def count_sequences_by_length(seqs, min_len):
count = 0
for seq in seqs:
if len(seq) >= min_len:
count += 1
return count
# 4) Number of sequences with GC content ≥ a given value, e.g. 50%. The GC content could be calculated as (num_of_G + num_of_C) / Total_num_of_AGCT.
def calculate_gc_content(seq):
return (seq.count('G') + seq.count('C')) / (seq.count('A') + seq.count('T') + seq.count('G') + seq.count('C')) * 100
def count_sequences_by_gc_content(seqs, min_gc):
count = 0
for seq in seqs:
if calculate_gc_content(seq) >= min_gc:
count += 1
return count
# 5) The combination of choices 3 and 4: Number of sequences with length ≥ a particular length and with GC content ≥ a particular value.
def count_sequences_by_length_and_gc(seqs, min_len, min_gc):
count = 0
for seq in seqs:
if len(seq) >= min_len and calculate_gc_content(seq) >= min_gc:
count += 1
return count
if __name__ == "__main__":
main()
=========================== Please select the sequences property that you want to display or press 0 to exit the program. 1) Total number of sequences 2) Number of pattern occurrences 3) Number of sequences with length >= min_len 4) Number of sequences with GC% >= min_GC 5) Number of sequences with length >= min_len and GC% >= min_GC Total number of sequences is 516. =========================== Please select the sequences property that you want to display or press 0 to exit the program. 1) Total number of sequences 2) Number of pattern occurrences 3) Number of sequences with length >= min_len 4) Number of sequences with GC% >= min_GC 5) Number of sequences with length >= min_len and GC% >= min_GC Number of occurrences of 'GGATC' is 357. =========================== Please select the sequences property that you want to display or press 0 to exit the program. 1) Total number of sequences 2) Number of pattern occurrences 3) Number of sequences with length >= min_len 4) Number of sequences with GC% >= min_GC 5) Number of sequences with length >= min_len and GC% >= min_GC Number of sequences with length >= min_len is 454. =========================== Please select the sequences property that you want to display or press 0 to exit the program. 1) Total number of sequences 2) Number of pattern occurrences 3) Number of sequences with length >= min_len 4) Number of sequences with GC% >= min_GC 5) Number of sequences with length >= min_len and GC% >= min_GC Num of seqs with gc >= min_gc is 258. =========================== Please select the sequences property that you want to display or press 0 to exit the program. 1) Total number of sequences 2) Number of pattern occurrences 3) Number of sequences with length >= min_len 4) Number of sequences with GC% >= min_GC 5) Number of sequences with length >= min_len and GC% >= min_GC Number of seqs with len >= min_len and gc >= min_gc is 11. =========================== Please select the sequences property that you want to display or press 0 to exit the program. 1) Total number of sequences 2) Number of pattern occurrences 3) Number of sequences with length >= min_len 4) Number of sequences with GC% >= min_GC 5) Number of sequences with length >= min_len and GC% >= min_GC Exiting...
Question3 (Average of tests)¶
Suppose test1.txt, test2.txt and test3.txt are results from three different experiments. The gene set used in each experiment is slightly different. Each row in the data file represents one gene, with the following format:
Accession_Number Number Number …
- First, load the data from all files to a single dictionary, with the accession number as the key and the reference to a list of data from all experiments as the value.
- Then calculate the average for each gene and store them in another dictionary.
- At the end, write this new dictionary to an output file called
test_averages.txt, where the first column is the accession number of the gene, and the second column is the corresponding average value.
def main():
files = ["test1.txt", "test2.txt", "test3.txt"]
new_dict = {}
for file in files:
with open(file, 'r') as f:
for line in f:
parts = line.strip().split()
gene = parts[0] # Set key
values = list(map(float, parts[1:])) # Set value
if gene not in new_dict:
new_dict[gene] = []
new_dict[gene].append(values)
test_averages = calculate_averages(new_dict)
with open("test_averages.txt", 'w') as f:
for gene, average in test_averages.items():
f.write(f"{gene}\t{average}\n")
print("Average of all genes have been written.")
# Calculate the average for each gene from different experiments.
def calculate_averages(data):
test_averages = {}
for gene, values in data.items():
total = 0
count = 0
for values_diffexp in values:
for value in values_diffexp:
total += value
count += 1
if count > 0:
test_averages[gene] = total / count
return test_averages
if __name__ == "__main__":
main()
Average of all genes have been written.
Question4 (Molecular Weight)¶
Write a program with the following steps.
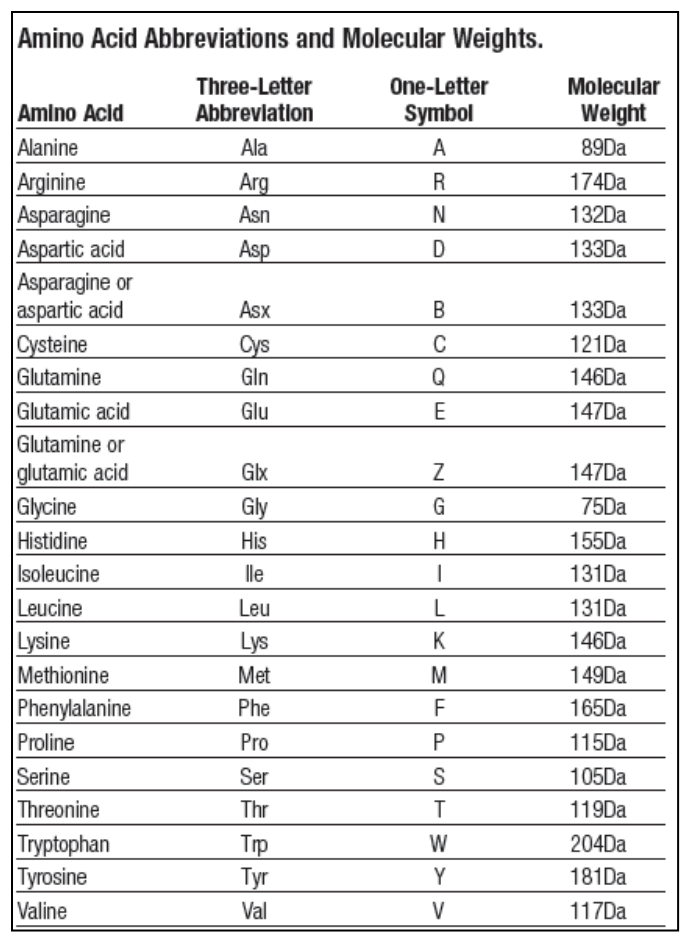
- Make a python dictionary of one-letter amino acids codes (the keys) to their molecular weight (the values), for all 22 amino acids. The molecular weight of 22 amino acids can be found in the table of next page. For example, the molecular weight of C (Cysteine) is 121.
- Print out a list of all the amino acids sorted by their molecular weights from the heaviest to the lightest. Hint: Just sort the items in Question (1) dictionary based on values. The output will look like:
AA MW W 204Da Y 181Da R 174Da F 165Da … … - Read the protein sequence from lysozyme.fasta and calculate the molecular weight of this protein using the dictionary created in question (1).
def main():
ac_dict = create_amino_acid_dict()
sort_amino_acids_by_weight(ac_dict)
protein_weight = calculate_protein_weight("lysozyme.fasta", ac_dict)
print(f"===========================\nThe molecular weight of the protein is: {protein_weight}Da")
# Make a python dictionary of one-letter amino acids codes (the keys) to their molecular weight (the values), for all 22 amino acids. T
def create_amino_acid_dict():
ac_dict = {'A': 89, 'R': 174, 'N': 132, 'D': 133, 'B': 133,
'C': 121, 'Q': 146, 'E': 147, 'Z': 147, 'G': 75,
'H': 155, 'I': 131, 'L': 131, 'K': 146, 'M': 149,
'F': 165, 'P': 115, 'S': 105, 'T': 119, 'W': 204,
'Y': 181, 'V': 117}
return ac_dict
# Print out a list of all the amino acids sorted by their molecular weights from the heaviest to the lightest.
def sort_amino_acids_by_weight(ac_dict):
sorted_amino_acids = sorted(ac_dict.items(), key=lambda item: item[1], reverse=True)
print("AA\tMW")
for amino_acid, weight in sorted_amino_acids:
print(f"{amino_acid}\t{weight}Da")
# Calculate the molecular weight of this `lysozyme.fasta` protein using the amino acids dictionary.
def calculate_protein_weight(filepath, ac_dict):
protein_sequence = ""
with open(filepath, "r") as file:
for line in file:
line = line.strip()
if line.startswith(">"):
continue # Skip the header line
protein_sequence += line
protein_weight = 0
for ac in protein_sequence:
if ac in ac_dict:
protein_weight += ac_dict[ac]
return protein_weight
if __name__ == "__main__":
main()
AA MW W 204Da Y 181Da R 174Da F 165Da H 155Da M 149Da E 147Da Z 147Da Q 146Da K 146Da D 133Da B 133Da N 132Da I 131Da L 131Da C 121Da T 119Da V 117Da P 115Da S 105Da A 89Da G 75Da =========================== The molecular weight of the protein is: 19421Da